Problems w/ power spectra?
Contents
Getting started
load SynthSig;
Aliasing
Here we need to generate another peak.
om_peak= 4.4*2*pi; ph = rand(1,1)*2*pi; sig.x_peak2 = 2*10.*cos(om_peak*sig.t+ph)/10; [p,f]=rawPowerSpec(sig.x_peak2+sig.x_peak+sig.x_w,dt); % Now, suppose that we can only sample at 0.25 Hz (i.e. 1/4 of the time) xx = sig.x_peak2+sig.x_peak+sig.x_w; x=xx(1:4:end); t = sig.t(1:4:end); [pn,fn]=rawPowerSpec(x,median(diff(t))); jmkfigure(100,2,0.6);clf subplot(2,1,1); plot(x(1:100)); hold on; plot(1:4:100,x(1:4:100),'b.','markersi',10); xlabel('t [s]'); ylabel('x [m]'); subplot(2,1,2); loglog(f,p,fn,pn) axis tight; yl = get(gca,'ylim'); xlabel('f [Hz]'); ylabel('\phi_x [m^2 s^{-1}]'); axis tight print -dpng -r250 doc/Aliasing
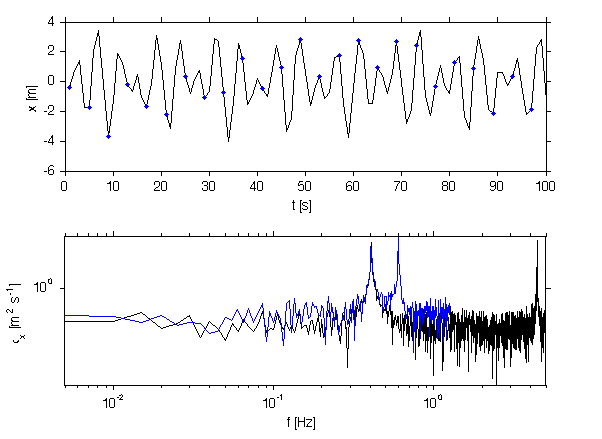
here we see the result of aliasing, where the peak has wrapped around to a harmonic in the resolved spectrum. To stop this we should smooth the new signal first:
[b,a]=butter(2,0.3); x = filtfilt(b,a,xx); subplot(2,1,1); plot(1:4:100,x(1:4:100),'r.','markersize',10) plot(x(1:100),'g') xlabel('t [s]'); ylabel('x [m]'); [pnp,fnp]=rawPowerSpec(x,median(diff(sig.t))); x = x(1:4:end); t = sig.t(1:4:end); [pnn,fnn]=rawPowerSpec(x,median(diff(t))); subplot(2,1,2); loglog(f,p,fn,pn,fnp,pnp,fnn,pnn) set(gca,'ylim',yl); xlabel('f [Hz]'); ylabel('\phi_x [m^2 s^{-1}]'); axis tight print -dpng -r250 doc/AliasingFixed
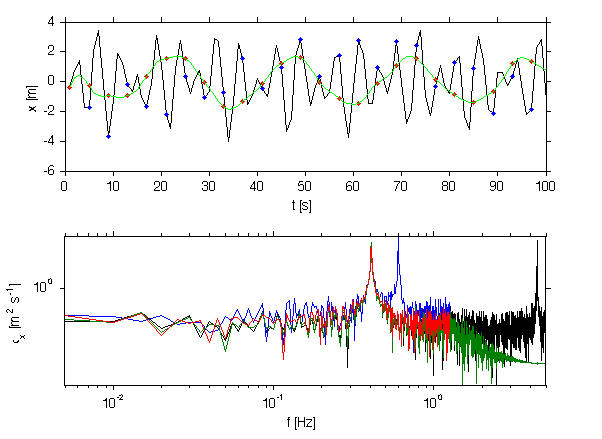
Finite record length
The last foible is finite record length. The difference between frequency bins is df = 1/T where T is the length of the record. Shorter records, the longer df.
To see the effect make a second peak very close to the first, but far enough apart that they can be distinguished in the full time-series.
om = th.f*2*pi; om_peak = find(om>0.4*2*pi); f0=om(om_peak(1))/2/pi; om_peak=om(om_peak(1)+3); % chose a peak 4 bins higher. ff = om_peak/2/pi; 1./abs(ff-f0) ph = rand(1,1)*2*pi; sig.x_peak2 = A_peak.*cos(om_peak*sig.t+ph)/10; x = sig.x_peak2+sig.x_peak+sig.x_w; jmkfigure(3,2,0.7);clf subplot(3,1,1); plot(x); hold on; plot(x(1:1500),'b'); plot(x(1:1000),'r'); ylabel('x [m]'); subplot(3,1,[2 3]); sty = {'k-','b-','r-'}; N = length(x); for i=1:3; [p,f]=rawPowerSpec(x(1:N-0.35*N*(i-1)),dt); loglog(f,p,sty{i}); hold on; h(i)=loglog(f,p,'o','markersi',6,'col',sty{i}(1)); set(gca,'xlim',[1.1*10^(-0.5) 1.7*10^(-0.5)]); df(i)=median(diff(f)); nn(i) = length(x(1:N-0.25*N*(i-1))); str{i}=sprintf('N=%d; \\Delta f=%1.2e [Hz]',nn(i),df(i)); end; xlabel('f [Hz]'); ylabel('\phi_x [m^2 Hz^{-1}]'); legend(h(1:3),str{1:3}) title(sprintf('Peaks separated by \\Delta f=%1.2e',abs(ff-f0))); print -dpng -r400 doc/FiniteRecord.jpg
ans = 66.6667
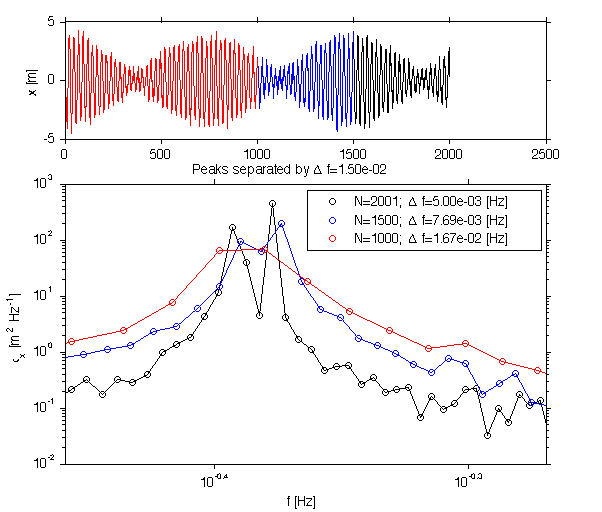
Here it is clear that fewer data points means poorer peak resolution. Once df rises above the peak separation they merge.
A trick to get better frequency resolution is to zero-pad the data. However, in this case, even if we do this, we do not improve resolution between the peaks, just the centering of the peaks.
jmkfigure(4,2,0.6);clf
sty = {'k-','b-','r-'};
N = length(x);
for i=1:3;
subplot3(1,3,i,'xscale','log','yscale','log');
[p,f]=rawPowerSpec(x(1:N-0.35*N*(i-1)),dt);
loglog(f,p,sty{i});
xx = x(1:N-0.35*N*(i-1));
xxx = zeros(1,floor(5*N));
xxx(1:length(xx)) =xx;
[pp,ff]=rawPowerSpec(xxx,dt);
h(i)=loglog(ff,pp,sty{i});
hold on;
h(i)=loglog(f,p,sty{i});
hold on;
% h(i)=loglog(f,p,'o','markersi',6,'col',sty{i}(1));
set(gca,'xlim',[1.1*10^(-0.5) 1.7*10^(-0.5)]);
df(i)=median(diff(f));
nn(i) = length(x(1:N-0.25*N*(i-1)));
str{i}=sprintf('N=%d',nn(i));
if i==1
ylabel('\phi_x [m^2 Hz^{-1}]');
end;
end;
%legend(h(1:3),str{1:3})
title('Zero-padded data');
xlabel('f [Hz]');
print -dpng -r400 doc/ZeroPad.jpg
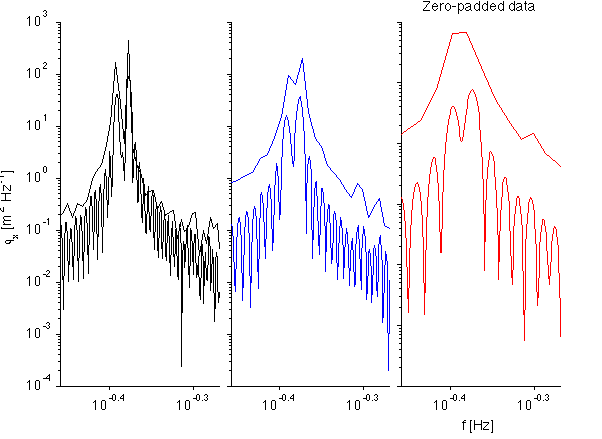
Note in this example how the poorly-resolved peaks (blue) have better peak localization than if we don't zero pad. However, this doesn't help separate peaks that aren't resolved in the unpadded timeseries.
Spectral Leakage:
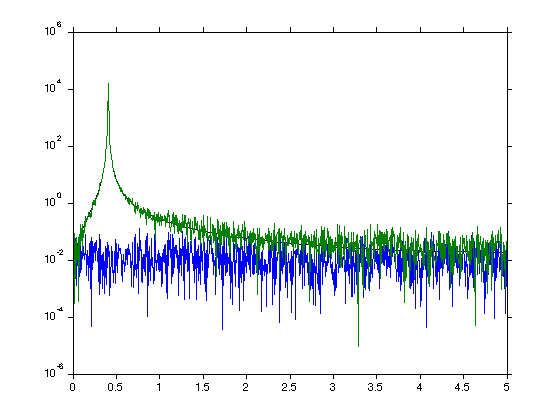
Compute a raw fft of a strong peak with white noise. Note that we have put the steps from the previous tutorial into a function.
dt = median(diff(sig.t)) [p,f]=rawPowerSpec(sig.x_peak*10,dt); [p_w,f]=rawPowerSpec(sig.x_w,dt); [p_p,f]=rawPowerSpec(sig.x_peak*10+sig.x_w,dt); figure(1);clf semilogy(f,p,f,p_w,f,p_p);
dt =
0.1000
Next
Next we consider ways to reduce smearing of peaks in the spectra and ways to improve the signal-to-noise of spectral extimates.